Here’s a heartfelt science story for all those Valentine’s day fans out there. Scientists from the Gladstone Institutes have identified how a group of transcription factors interact during embryonic development to make a healthy heart. Their work will increase our biological understanding of how the heart is formed and could produce new methods for treating cardiovascular disease.
The study, published today in the journal Cell, describes a tumultuous love story between cardiac transcription factors. Transcription factors are proteins that orchestrate gene expression. They have the power to turn genes on or off by binding to specific DNA sequences and recruiting other proteins that will eventually turn the information encoded in that gene into a functional protein.
Every organ has its own special group of transcription factors that coordinate the gene expression required for that organ’s development. Often times, transcription factors within a group directly interact with each other and work together to conduct a specific sequence of events. These interactions are essential for making healthy tissues and organs, but scientists don’t always understand how these interactions work.
For the heart, scientists have already identified a group of transcription factors essential for cardiac development, and genetic mutations in any of these factors can impair heart formation and cause heart defects in newborns. What’s not known, however, are the details on how some of these cardiac transcription factors interact to get their job done.
A cardiac love triangle
In the Gladstone study, the scientists focused on how three key cardiac transcription factors – NKX2.5, TBX5, and GATA4 – interact during heart development. They first proved that these transcription factors are essential for the formation of the heart in mouse embryos. When they eliminated the presence of one of the three factors from the developing mouse embryo, they observed abnormal heart development and heart defects. When they removed two factors (NKX2.5 and TBX5), the results were even worse – the heart wasn’t able to form and none of the embryos survived.
Next, they studied how these transcription factors interact to coordinate gene expression in heart cells called cardiomyocytes made from mouse embryonic stem cells that lacked either NKX2.5, TBX5, or both of these factors. Compared to normal heart cells, cardiomyocytes that lacked one or both of these two transcription factors started beating at inappropriate times – either earlier or later than the normal heart cells.
Taking a closer look, the scientists discovered that TBX5, NKX2.5 and GATA4 all hangout in the same areas of the genome in embryonic stem cells that are transitioning into cardiomyocytes. In fact, each individual transcription factor required the presence of the others to bind their DNA targets. If one of these factors was missing and the love triangle was broken, the remaining transcription factors became confused and bound random DNA sequences in the genome, causing a mess by turning on genes that shouldn’t be on.
First author on the study, Luis Luna-Zurita, explained the importance of maintaining this cardiac love triangle in a Gladstone Press Release:
“Transcription factors have to stick together, or else the other one goes and gets into trouble. Not only are these transcription factors vital for turning on certain genes, but their interaction is important to keep each other from going to the wrong place and turning on a set of genes that doesn’t belong in a heart cell.”
Crystal structure tells all
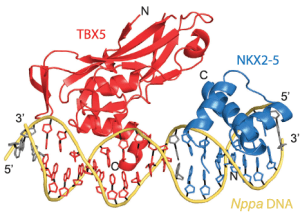
Protein crystal structure of NKX2.5 and TBX5 bound to DNA. (Luna-Zurita et al. 2016)
The last part of the study proved that two of these factors, NKX2.5 and TBX5, directly interact and physically touch each other when they bind their DNA targets. In collaboration with a group from the European Molecular Biology Laboratory (EMBL) in Germany, they developed protein crystal structures to model the molecular structure of these transcription factors when they bind DNA.
Co-author and EMBL scientist Christoph Muller explained his findings:
“The crystal structure critically shows the interaction between two of the transcription factors and how they influence one another’s binding to a specific stretch of DNA. Our detailed structural analysis revealed a direct physical connection between TBX5 and NKX2-5 and demonstrated that DNA plays an active role in mediating the interaction between the two proteins.”
Big picture
While this study falls in the discovery research category, its findings increase our understanding of the steps required to make a healthy heart and sheds light on what goes wrong in patients or newborns with heart disease.
Senior author on the paper and Gladstone Professor Benoit Bruneau explained the biomedical applications of their study for treating human disease:
“Gene mutations that cause congenital heart disease lower the levels of these transcription factors by half, and we’ve shown that the dosage of these factors determines which genes are turned on or off in a cell. Other genetic variants that cause heart defects like arrhythmias also affect the function of these factors. Therefore, the better we understand these transcription factors, the closer we’ll come to a treatment for heart disease. Our colleagues at Gladstone are using this knowledge to search for small molecules that can affect gene regulation and reverse some of the problems caused by the loss of these transcription factors.”
I think it’s worth mentioning that these studies were done using mouse embryos and mouse embryonic stem cells. Future work should be done to determine whether this cardiac love triangle and the same transcription factor interactions exist in human heart cells.
Related Links:


